The Microbial Super-Resolution Microscopy Lab


Current Projects
For Publications see here or Google scholar

From left to right: UPEC invades target cells. Intracellular bacterial communities (IBCs) are formed and multiply into great numbers. When the cells cannot cope with the IBCs, they rupture and filamentous UPEC are readily observed. The molecular details of the late stages of this process and how filaments revert back to rods are poorly understood. Specifically, we want to know the state of the cell division machinery during various stages of filamentation and reversal.

UPEC (magenta) expressing FtsZ-mNG in the process
of infecting bladder cells (blue membrane, grey nucleus).
Understanding bacterial morphology changes during Urinary Tract Infections
Urinary tract infections (UTIs) are a global health issue that affects millions of people each year.
We aim to identify molecular mechanisms that govern division and filamentation of UPEC* cells during infection of human bladder cells. We are looking at the assembly dynamics and composition of the cell division machinery during various stages of the infection cycle. One specific aim is to look at a special class of proteins called SPOR-domain proteins, E. coli has four: FtsN, DamX, DedD, and RlpA. One of these, DamX, has been shown to be essential for filamentation. The goal is to better understand the roles of all SPOR-domain proteins during UTI, as they may hold the key for novel therapeutics.
We are also looking at interspecies bacterial behaviours in multispecies infection models using a combination of five clinically relevant UTI pathogens: UPEC, Enterococcus Faecalis, Klebsiella pneumoniae, Pseudomonas aeruginosa and Proteus mirabilis.
* Uropathogenic Escherichia coli,
the most common causative agent of UTI.
A UPEC filament that initially grows before reverting back to rods after infection.
Cells are labelled with sfGFP.
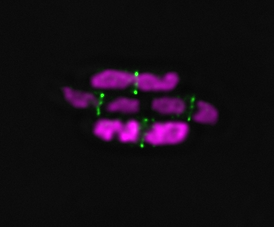
2 color STED on DNA and FtsN
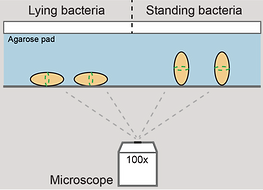
Schematic view of standing cells

2 color STED showing the organization of FtsZ and FtsN in a standing cell

PALM imaging of division protein DamX in cells in a standing position
High-resolution imaging of the bacterial cell division machinery
The goal of this project is literally to get a better picture of the cell division machinery in bacteria. Cell division in E. coli is guided by the protein FtsZ, that together with ~ 30 additional proteins work together to divide the cells.
We are currently developing protocols to use uropathogenic E. coli (UPEC) as a model system. Combining the best of multicolour super-resolution fluorescence microscopy, microfluidics and nano-engineering are we obtaining new high-resolution information about the internal organisation of the components of the division machinery.
The overall rationale behind this is:
"If we know how bacteria divide, can we perhaps figure out how to stop them from doing so, and in that way stop infection?"
E. coli cells growing and dividing in a "mother machine".
Green: Division machinery (FtsZ-mVenus).
Red: DNA (HupA-RFP)
Developing Forensic Microbiology in an Australian context
Accurately determining time since death of decomposing human remains is hard: a far cry from how the TV shows depict it!
We are exploring ways to utilize the rapid changes in the microbial communities after death to increase the accuracy of time since death estimations. This is possible because many bacteria operate at a faster time scale than humans, which results in changes to the overall bacterial community composition taking place over hours rather than days or weeks. These time-depended microbial community changes have been given the name the ‘microbial clock’. With this research, we also learn about the fundamental roles microbes play in the human decomposition process, particularly in an Australian setting.
Working together with the UTS Forensics and Chemistry teams, we are developing approaches based on genomic sequencing and optical imaging to follow changes during the decomposition process of human remains as seen from a microbiological viewpoint. We mainly do this work in the field, at the Australian Facility for Taphonomic Experimental Research (AFTER), located in the north-west of Sydney, the only facility in Australia (and the southern hemisphere) dedicated to research on various aspects of human post-mortem and decomposition investigations.
The long-term goal is to create a tool based on the microbial clock that will increase the capabilities of Australian law enforcements agencies to better pin-point the post-mortem interval, a critically important aspect of crime investigations.

Cell shape and morphology
As a side project do we also look at how the cell division machinery is behaving in artificially remodelled cells. These cells are shaped into Hearts, Stars, Triangles ect. and imaged by super-resolution microscopy.
The aim is to figure out to what extent cell shape influences the division process.
Live E. coli cells were shaped into various geometrical forms. The cell division protein FtsZ was labeled with mCitrine or mNeonGreen and image by STED nanoscopy. Pseudo-coloured image.

Current funding comes from:

